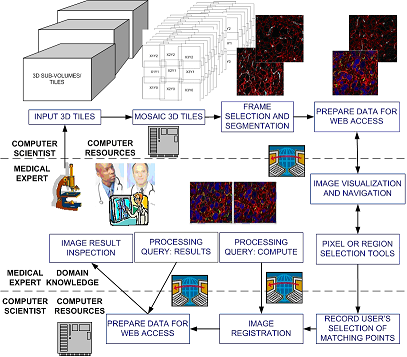
Figure 1: An overview of the current prototype.
In a collaborative environment with medical and computer science collaborators, the goal is to reconstruct 3D medical volume from high resolution microscopy images of several cross sections. High resolution mosaic images of cross sections are formed from a large set of tiles, and then the mosaic images are aligned to construct a 3D volume.
From the medical collaborator viewpoint, 3D volume reconstruction requires too much computation to be handled by a desktop computer. From the computer science collaborator viewpoint, 3D volume reconstruction requires selecting pairs of matching features for cross section alignment and hence medical expertise. Thus, there is need to develop a cyber-infrastructure environment where the computational resources and the expertise of remotely located medical and computer science collaborators can be integrated. We proposed to use web services for building such a cyber-infrastructure environment, and developed a prototype system that enables researchers from UIC and NCSA UIUC to collaborate. The overview of the current prototype is in Figure 1.
The proposed solution consists of the following workflow. First, the medical collaborator acquires images and sends data to the computer science collaborator. It is also possible that the medical collaborator uploads the data assuming a high bandwidth connection. Second, a computer program automatically mosaics image tiles, selects the most salient frame from each sub-volume, segments the selected frames and pre-computes centroids of all segments. The pre-processed images and centroid information are packaged for web access. Third, the medical collaborator will be notified about the URL designed for accessing and navigating the image data, as well as for selecting registration points and visualizing registration results. The medical collaborator selects matching features for cross section alignment by using standard human computer interfaces (HCI), and our developed image navigation tools. The points are saved at NCSA UIUC for additional processing after pressing the button “Compute". Pressing this button sends a query from UIC to NCSA to request registration computation. After the computation is completed, the results can be displayed by pressing the button “Result”. Pressing the button “Result” executes (a) a query transmission from UIC to NCSA, (b) image transformation according to the computed parameters, and (c) a transmission of the resulting image back to UIC. In the aforementioned workflow, all operations that require intensive computation are performed at NCSA (computer resource location), while all operations that require medical domain knowledge (image acquisition, registration point selection, and volume inspection) are performed at UIC (domain expertise location).

Figure 1: An overview of the current prototype.