We are interested in developing a framework for three-dimensional volume reconstruction. 3D volume reconstruction is currently missing from many medical data analysis techniques. The problem is challenging due to significant variations of intensity and shape of cross sectioned structures, unpredictable and inhomogeneous geometrical warping during medical specimen preparation, and an absence of external fiduciary markers. The framework addresses the problem of automation in the presence of the above challenges as they are frequently encountered during 3D volume reconstructions used for cell biology and medical investigations.
The goal is to reconstruct 3D volume from high resolution microscopy images of several images or cross sections. They are formed from a large set of tiles, and then the mosaic images are aligned to construct a 3D volume. Reconstruction usually requires too much computation to be handled by a desktop computer. Additionally, in our work, the three-dimensional (sub) volume registration problem is viewed primarily as an alignment problem. It is generally approached by extracting two- or three-dimensional features from each sub-volume and registering the sub-volumes based on the analysis of detected features.
Potential applications include:
- Processing and Analysis of Microscopy Imagery (Confocal, MRI etc.)
- Microarray Data Analysis
- Novel Imaging in Medicine
3D Medical Volume Reconstruction for CLSM
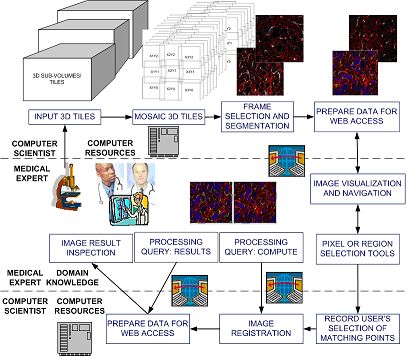
The overview of the current cyber-infrastructure environment prototype.
We present a problem of three-dimensional volume reconstruction from fluorescent confocal laser scanning microscopy (CLSM) imagery. We overview a three-dimensional volume reconstruction framework which consists of:
- volume reconstruction procedures using multiple automation levels, feature types, and feature dimensionalities,
- a data-driven registration decision support system,
- an evaluation study of registration accuracy, and
- a novel intensity enhancement technique for 3D CLSM volumes.
The motivation for developing the framework came from the lack of 3D volume reconstruction techniques for CLSM image modality. The 3D volume reconstruction problem is challenging due to significant variations of intensity and shape of cross sectioned structures, unpredictable and inhomogeneous geometrical warping during medical specimen preparation, and an absence of external fiduciary markers. The framework addresses the problem of automation in the presence of the above challenges as they are frequently encountered during CLSM-based 3D volume reconstructions used for cell biology investigations.
There is need to develop a cyber-infrastructure environment where the computational resources and the expertise of remotely located medical and computer science collaborators can be integrated. We proposed to use web services for building such a cyber-infrastructure environment, and developed a prototype system that enables researchers to collaborate, in our case researchers from NCSA and Department of Pathology, University of Illinois at Chicago.
Tools for registration and alignment (part of the CLMS project):
Alignment Tool 1:
Transformation Selection (Blood vessels)
Alignment Tool 2: Image Size Selection
This project was supported by the National Institute of Health and National Center for Supercomputing Applications.
Integration, Analysis and Visualization of Large Size Multi-Modal 3D Medical Imagery
Large size datasets from Magnetic resonance imaging (MRI) microscopy and 3D diffusion tensor (DTI) weighted fiber tracking imagery are integrated to produce the neuron-anatomical model of song bird. A zebra finch (Taeniopygia guttata) is an ideal animal model since the auditory behavior, the neural pathways, the genomic profile, and the neural pathways for sound generation are similar to humans.
The joint research of NCSA and College of Applied Life Studies (ALS) combines the computer science and bio-medical expertise not only to understand neuron-anatomical model but also to test hypotheses about efficient diagnoses from multi-modal imagery. High resolution MRI images of the zebra finch brain and 3D diffusion tensor weighted fiber tracking imaging were fused with serial cyto-histological sections of the complete brain of a male and female zebra finch.
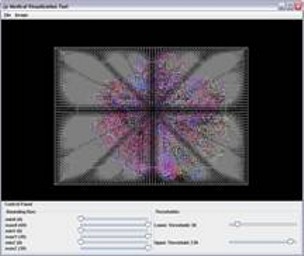
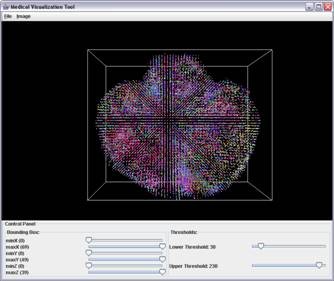
Three dimensional visualization of Color Coded diffusion tensor imaging (DTI) data of the bird's brain with significant background noise (first image). The DTI data is integrated with the corresponding magnetic resonance (MRI) image (not shown here). Using MRI dataset as a 3D mask the noise is reduced and final 3D DTI image is computed (second image).
DNA Microarray data processing
Microarray grid alignment and foreground separation are the basic processing steps of DNA microarray images that affect the quality of gene expression information, and hence impact our confidence in any data-derived biological conclusions. Thus, understanding microarray data processing steps becomes critical for performing optimal microarray data analysis.
The workflow of microarray data processing starts with raw image data acquired with laser scanners and ends with the results of data mining that have to be interpreted by biologists. The microarray data processing workflow includes issues related to (1) data management (e.g., MIAME compliant database, (2) image processing (grid alignment, foreground separation, spot quality assessment, data quantification and normalization, (3) data analysis (identification of differentially expressed genes, data mining, integration with other knowledge sources, and quality and repeatability assessments of results, and (4) biological interpretation (visualization). The main objective of this project is related to image processing, namely grid alignment, foreground separation, spot quality assessment, data quantification, normalization and visualization.
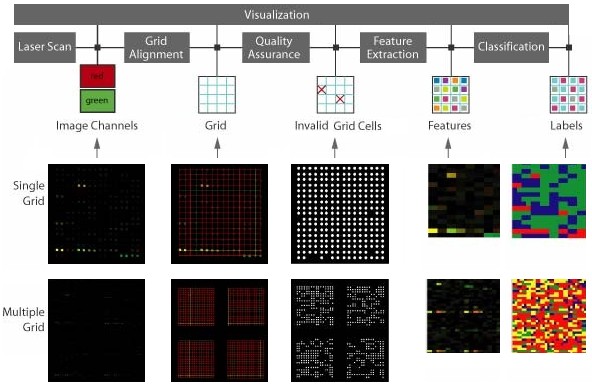
Microarray data processing workflow: Fluorescent DNA microarray images obtained from laser scanners containing a 2D array of dots with two channels of 532nm (red) and 632nm (green) wavelengths. The grid alignment is performed producing a set of lines intersecting at each dot. Dots define a valid foreground. Quality assurance screening eliminates grid cells with unreliable microarray information. Finally, image of sample mean values extracted at each grid cell using particular mask is extracted and colored in a red-green-blue space with color assigned to each cluster/pixel. Statistics of each cluster can be viewed in the text area.
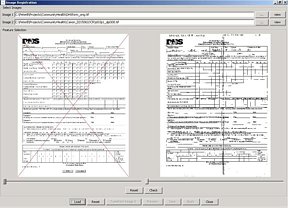
Registration tool. (Left) Original CHEC form and faxed and scanned copy (right).
ECHO: Electronic Children Health Observatory
The purpose of our project is to develop and pilot a state-wide, state-of-the-art student personal health record tracking system called ECHO (Electronic Children Health Observatory) that can screen and track childhood obesity, as well as provide intervention if needed, for the State of Illinois.
There are three specific aims in this study: (1) To complete the development of the major system architecture of ECHO and its application elements, (2) To pilot the system in different application settings and to evaluate the efficiency and utility of the system, (3) To modify the system based on the feedback, develop instructions for the future implementation of the system and estimate the cost of the implementation.